如何使用知识图谱和矢量数据库实现Graph RAG
在本篇文章中,我将重点介绍一种将知识图谱和 LLM 结合起来使用的流行方法: 使用知识图谱的检索增强生成(RAG),有时也称为图谱检索增强生成(Graph RAG)、检索增强生成(GraphRAG)、检索增强生成(GRAG)或语义检索增强生成(Semantic RAG)。检索增强生成(RAG)是通过检索相关信息来增强发送给 LLM 的提示,从而生成响应。我们的想法是,与其将提示信息直接发送给没有经过数据训练的 LLM,不如在提示信息中添加 LLM 所需的相关信息,以便 LLM 准确回答你的提示信息。由于知识图谱是为存储知识而构建的,因此它是存储内部数据和为 LLM 提示补充额外上下文的完美方式,从而提高了回复的准确性和上下文理解能力。
重要的是,我认为经常被误解的是,RAG 和使用 KG 的 RAG(图形 RAG)是结合技术的方法,而不是产品或技术本身。没有人发明、拥有或垄断 Graph RAG。不过,大多数人都能看到这两种技术结合后的潜力,而且越来越多的研究证明了这两种技术结合的好处。
一般来说,在 RAG 的检索部分使用 KG 有三种方法:
- 基于矢量的检索: 将 KG 矢量化并存储在矢量数据库中。如果再将自然语言提示矢量化,就可以在矢量数据库中找到与提示最相似的矢量。由于这些向量与图中的实体相对应,因此你可以根据自然语言提示返回图中最 “相关 ”的实体。请注意,你可以在没有图的情况下进行基于向量的检索。这实际上是 RAG 最初的实现方式,有时也称为基准 RAG。你可以将 SQL 数据库或内容矢量化,然后在查询时进行检索。
- 提示到查询检索: 使用 LLM 为你编写 SPARQL 或 Cypher 查询,针对你的 KG 使用该查询,然后使用返回的结果来增强你的提示。
- 混合(向量 + SPARQL): 你可以将这两种方法以各种有趣的方式结合起来。在本教程中,我将演示结合这两种方法的一些方法。我将主要侧重于使用矢量化进行初始检索,然后使用 SPARQL 查询来完善结果。
不过,有很多方法可以将向量数据库和 KG 结合起来进行搜索、相似性分析和 RAG 分析。这只是一个说明性的例子,旨在强调每种方法各自的优缺点以及将它们结合起来使用的好处。我在这里将它们结合起来使用的方式--矢量化用于初始检索,然后 SPARQL 用于过滤--并不是独一无二的。我在其他地方也看到过这种方法。我在一家大型家具制造商那里听说过一个很好的例子。他说,矢量数据库可能会向购买沙发的人推荐绒毛刷,但知识图谱会了解材料、属性和关系,并确保不会向购买皮沙发的人推荐绒毛刷。
在本文中,我将:
- 将数据集矢量化为矢量数据库,以测试语义搜索、相似性搜索和 RAG(基于矢量的检索)
- 将数据转化为一个 KG,以测试语义搜索、相似性搜索和 RAG(提示到查询检索,但实际上更像是查询检索,因为我只是直接使用 SPARQL,而不是让 LLM 将我的自然语言提示转化为 SPARQL 查询)
- 将带有知识图谱中的标签和 URI 的数据集矢量化为矢量数据库(我将称之为 “矢量化知识图谱”),并测试语义搜索、相似性和 RAG(混合)。
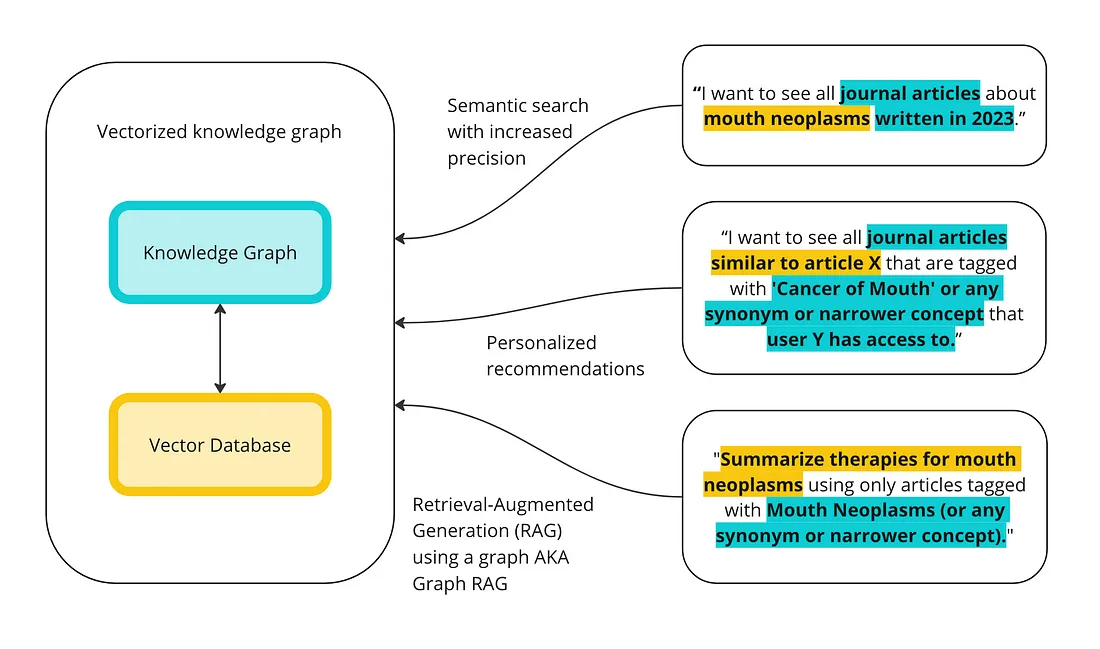
目的是说明知识图谱和矢量数据库在这些功能上的区别,并展示它们可以协同工作的一些方式。以下是矢量数据库和知识图谱如何共同执行高级查询的高级概述。
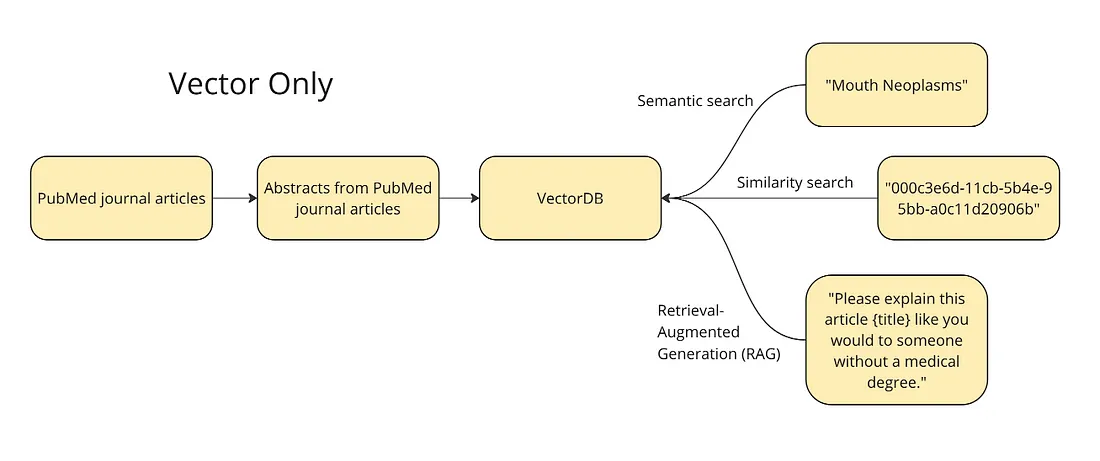
第1步:基于向量的检索
下图显示了该计划的总体情况。我们希望将期刊论文的摘要和标题矢量化到一个矢量数据库中,以运行不同的查询:语义搜索、相似性搜索和简单版本的 RAG。对于语义搜索,我们将测试 “口腔肿瘤 ”这样的术语--矢量数据库应返回与该主题相关的文章。对于相似性搜索,我们将使用给定文章的 ID 来查找其在向量空间中的近邻,即与这篇文章最相似的文章。最后,矢量数据库允许一种 RAG 形式,我们可以用一篇文章来补充类似 “请向没有医学学位的人解释一下 ”这样的提示。
我决定使用 PubMed 资料库中的 50,000 篇研究文章数据集(许可证 CC0:公共领域)。该数据集包含文章标题、摘要以及元数据标签字段。这些标签来自医学主题词表(MeSH)控制词汇词库。在本部分教程中,我们只使用摘要和标题。这是因为我们要比较的是矢量数据库和知识图谱,而矢量数据库的优势在于它能够 “理解 ”没有丰富元数据的非结构化数据。我只使用了数据的前 10,000 行,只是为了让计算运行得更快。
下面是 Weaviate 的官方快速入门教程。
from weaviate.util import generate_uuid5
import weaviate
import json
import pandas as pd
#Read in the pubmed data
df = pd.read_csv("PubMed Multi Label Text Classification Dataset Processed.csv")
然后,我们就可以建立与 Weaviate 集群的连接:
client = weaviate.Client(
url = "XXX", # Replace with your Weaviate endpoint"XXX", # Replace with your Weaviate endpoint
auth_client_secret=weaviate.auth.AuthApiKey(api_key="XXX"), # Replace with your Weaviate instance API key
additional_headers = {
"X-OpenAI-Api-Key": "XXX" # Replace with your inference API key
}
)
在将数据矢量化到矢量数据库之前,我们必须定义模式。在这里,我们要定义要矢量化的 csv 列。如前所述,就本教程而言,开始时我只想矢量化标题和摘要列。
class_obj = {= {
# Class definition
"class": "articles",
# Property definitions
"properties": [
{
"name": "title",
"dataType": ["text"],
},
{
"name": "abstractText",
"dataType": ["text"],
},
],
# Specify a vectorizer
"vectorizer": "text2vec-openai",
# Module settings
"moduleConfig": {
"text2vec-openai": {
"vectorizeClassName": True,
"model": "ada",
"modelVersion": "002",
"type": "text"
},
"qna-openai": {
"model": "gpt-3.5-turbo-instruct"
},
"generative-openai": {
"model": "gpt-3.5-turbo"
}
},
}然后,我们将此模式推送到 Weaviate 集群:
client.schema.create_class(class_obj)schema.create_class(class_obj)
你可以直接在 Weaviate 集群中查看是否成功。
现在我们已经建立了模式,可以将所有数据写入矢量数据库。
import logging
import numpy as np
# Configure logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s %(levelname)s %(message)s')
# Replace infinity values with NaN and then fill NaN values
df.replace([np.inf, -np.inf], np.nan, inplace=True)
df.fillna('', inplace=True)
# Convert columns to string type
df['Title'] = df['Title'].astype(str)
df['abstractText'] = df['abstractText'].astype(str)
# Log the data types
logging.info(f"Title column type: {df['Title'].dtype}")
logging.info(f"abstractText column type: {df['abstractText'].dtype}")
with client.batch(
batch_size=10, # Specify batch size
num_workers=2, # Parallelize the process
) as batch:
for index, row in df.iterrows():
try:
question_object = {
"title": row.Title,
"abstractText": row.abstractText,
}
batch.add_data_object(
question_object,
class_name="articles",
uuid=generate_uuid5(question_object)
)
except Exception as e:
logging.error(f"Error processing row {index}: {e}")
要检查数据是否已进入群集,可以运行以下命令:
client.query.aggregate("articles").with_meta_count().do().query.aggregate("articles").with_meta_count().do()由于某些原因,我只有 9997 行被矢量化。
使用向量数据库进行语义搜索
当我们谈到矢量数据库中的语义时,我们指的是使用LLM API将术语矢量化到矢量空间中,而LLM API是在大量非结构化内容上经过训练的。这意味着,向量会将术语的上下文考虑在内。例如,如果在训练数据中,马克-吐温(Mark Twain)与塞缪尔-克莱门斯(Samuel Clemens)被多次提及,那么这两个术语的向量在向量空间中应该彼此接近。同样,如果在训练数据中多次出现 Mouth Cancer 一词和 Mouth Neoplasms 一词,我们预计有关 Mouth Cancer 的文章的向量在向量空间中应该靠近有关 Mouth Neoplasms 的文章。
你可以运行一个简单的查询来检查它是否有效:
response = (
client.query
.get("articles", ["title","abstractText"])get("articles", ["title","abstractText"])
.with_additional(["id"])
.with_near_text({"concepts": ["Mouth Neoplasms"]})
.with_limit(10)
.do()
)
print(json.dumps(response, indent=4))
结果如下:
- 文章1:“牙龈转移是上皮样恶性间皮瘤多器官播散的先兆”。这篇文章讲述的是一项针对恶性间皮瘤(肺癌的一种)扩散到牙龈的患者进行的研究。这项研究的目的是测试不同治疗方法(化疗、脱屑治疗和放疗)对癌症的影响。这似乎是一篇合适的文章--它是关于牙龈肿瘤的,牙龈肿瘤是口腔肿瘤的一个分支。
- 文章2:"小唾液腺源性肌上皮瘤。光镜和电子显微镜研究"。这篇文章讲述的是从一名 14 岁男孩的牙龈上切除的肿瘤,肿瘤已经扩散到上颚的一部分,肿瘤由起源于唾液腺的细胞组成。这似乎也是一篇合适的文章--它讲述的是从一个男孩的口腔中切除的肿瘤。
- 文章3:"下颌骨转移性神经母细胞瘤。病例报告"。这篇文章是关于一名下颚患癌的 5 岁男孩的病例研究。这是一篇关于癌症的文章,但严格来说并非口腔癌--下颌骨肿瘤(下颌骨肿瘤)并非口腔肿瘤的子集。
这就是我们所说的语义搜索--这些文章的标题或摘要中都没有 “口腔 ”一词。第一篇文章是关于牙龈(牙龈)肿瘤的,它是口腔肿瘤的一个子集。第二篇文章是关于起源于受试者唾液腺的牙龈肿瘤,这两种肿瘤都是口腔肿瘤的子集。第三篇文章是关于下颌骨肿瘤--严格来说,根据 MeSH 词汇表,下颌骨肿瘤并不是口腔肿瘤的子集。不过,矢量数据库知道下颌骨与口腔很接近。
使用矢量数据库进行相似性搜索
我们还可以使用矢量数据库查找相似文章。我选择了一篇使用上述口腔肿瘤查询返回的文章,标题为 “牙龈转移是上皮样恶性间皮瘤多器官播散的首发症状”。利用这篇文章的 ID,我可以在矢量数据库中查询到所有类似的实体:
response = (
client.query
.get("articles", ["title", "abstractText"])"articles", ["title", "abstractText"])
.with_near_object({
"id": "a7690f03-66b9-5d17-b765-8c6eb21f99c8" #id for a given article
})
.with_limit(10)
.with_additional(["distance"])
.do()
)
print(json.dumps(response, indent=2))
结果按相似度排序。相似度以向量空间中的距离计算。如图所示,排在最前面的是牙龈文章--这篇文章是与自己最相似的文章。
其他文章是:
- 文章4:“针对烟草使用者进行口腔恶性病变筛查的可行性研究”。这篇文章是关于口腔癌的,但讲的是如何让吸烟者报名参加筛查,而不是他们的治疗方法。
- 文章5:“针对恶性胸膜间皮瘤的扩展胸膜切除术和去皮层术是一种对老年人有效且安全的去细胞手术”。这篇文章介绍了一项关于用胸膜切除术和去皮层术(从肺部切除癌症的手术)治疗老年人胸膜间皮瘤(肺癌)的研究。因此,这篇文章的类似之处在于它是关于间皮瘤的治疗方法,而不是牙龈肿瘤。
- 文章6: “下颌骨转移性神经母细胞瘤。病例报告"。同样,这篇文章是关于一名下颚患癌的 5 岁男孩的。这篇文章是关于癌症的,但严格来说不是口腔癌,也不像牙龈文章那样是关于治疗效果的。
可以说,所有这些文章都与我们最初的牙龈文章相似。很难评估它们有多相似,因此也很难评估相似性搜索的效果如何,因为这在很大程度上取决于用户对 “相似 ”的理解。你是否对有关间皮瘤治疗方法的其他文章感兴趣,而第一篇文章是关于间皮瘤如何扩散到牙龈的这一事实并不重要?在这种情况下,第 5 条是最相似的。或者,你是否对通过治疗或预防减少任何类型的口腔癌感兴趣?在这种情况下,第 4 条最为相似。矢量数据库的一个缺点是它是一个黑盒子--我们不知道为什么会返回这些文章。
使用矢量数据库进行检索增强生成(RAG)
下面是如何使用矢量数据库检索结果,然后将结果发送到 LLM 进行汇总--这是 RAG 的一个示例。
response = (
client.query
.get("articles", ["title", "abstractText"])get("articles", ["title", "abstractText"])
.with_near_text({"concepts": ["Gingival metastasis as first sign of multiorgan dissemination of epithelioid malignant mesothelioma"]})
.with_generate(single_prompt="Please explain this article {title} like you would to someone without a medical degree.")
.with_limit(1)
.do()
)
print(json.dumps(response, indent=4))
请看下面的回复:
“当然!这篇文章讲述的是一个人罹患一种叫做上皮样恶性间皮瘤的癌症的病例。这种癌症通常起病于肺部或腹部内膜。然而,在这个病例中,癌症扩散到身体其他部位的第一个迹象出现在牙龈(齿龈)上。转移是指癌细胞从原来的肿瘤扩散到身体的其他部位。在这个病例中,癌细胞在扩散到其他器官之前已经扩散到了牙龈。总之,这篇文章强调了早期发现和监测癌症的重要性,以及癌症扩散到身体意外部位的可能性。
事实上,我对这一回应感到失望。摘要清楚地解释了这是一项跟踪 13 名转移性恶性间皮瘤患者接受不同治疗及其结果的研究。RAG 输出将这篇文章描述为 “一个人”,完全没有提及这项研究。
与其只总结一篇文章,不如试着总结几篇文章。在下一个例子中,我们使用与上面相同的搜索词(口腔肿瘤),然后发送前三篇文章,并提示 "用要点概括这里的关键信息。让没有医学学位的人也能看懂",发送给一位法学硕士。
response = (
client.query
.get(collection_name, ["title", "abstractText"])get(collection_name, ["title", "abstractText"])
.with_near_text({"concepts": ["Mouth Neoplasms"]})
.with_limit(3)
.with_generate(grouped_task="Summarize the key information here in bullet points. Make it understandable to someone without a medical degree.")
.do()
)
print(response["data"]["Get"]["Articles"][0]["_additional"]["generate"]["groupedResult"])
这里是结果:
- Metastatic malignant mesothelioma to the oral cavity is rare, with more cases in jaw bones than soft tissue
- Average survival rate for this type of cancer is 9-12 months
- Study of 13 patients who underwent neoadjuvant chemotherapy and surgery showed a median survival of 11 months
- One patient had a gingival mass as the first sign of multiorgan recurrence of mesothelioma
- Biopsy of new growing lesions, even in uncommon sites, is important for patients with a history of mesothelioma
- Myoepithelioma of minor salivary gland origin can show features indicative of malignant potential
- Metastatic neuroblastoma in the mandible is very rare and can present with osteolytic jaw defects and looseness of deciduous molars in children
在我看来,这比之前的答复更好--它提到了第 1 条中进行的研究、治疗方法和结果。倒数第二项是关于 "小唾液腺源性肌上皮瘤。光镜和电子显微镜研究 "一文,似乎是准确的一行描述。最后一项是关于上文提到的第 3 条,也似乎是准确的一行描述。
第2步:使用知识图谱进行数据检索
以下是我们如何使用知识图谱进行语义搜索、相似性搜索和 RAG 的高级概述:
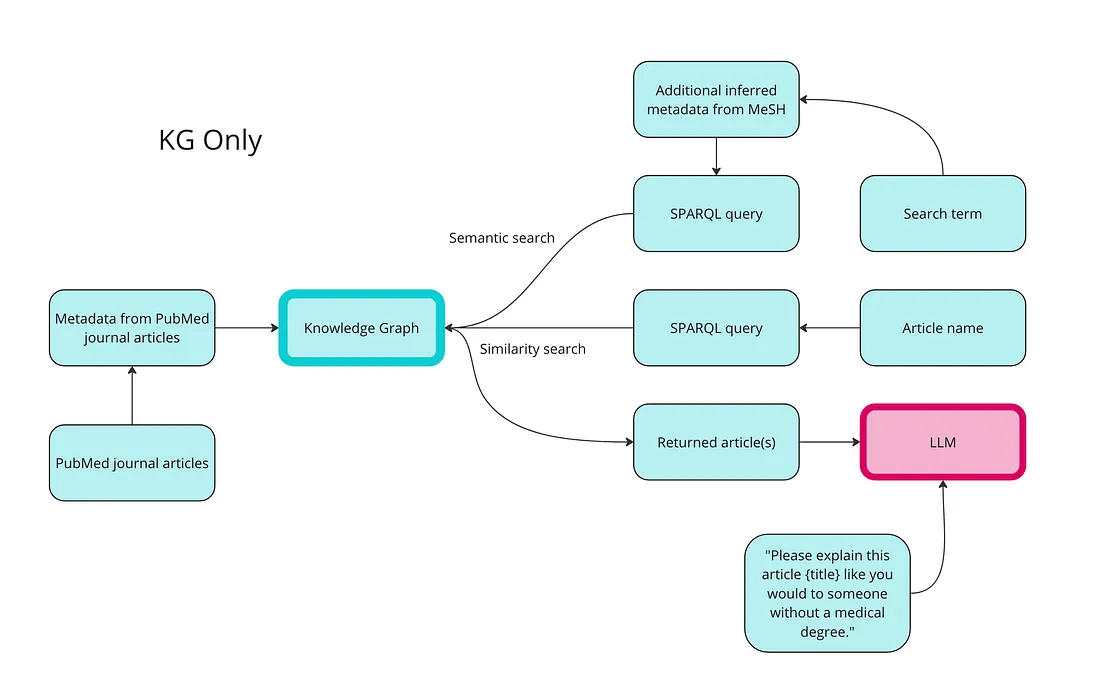
使用知识图谱检索数据的第一步是将数据转换成 RDF 格式。下面的代码为所有数据类型创建了类和属性,然后用文章和 MeSH 术语的实例填充。我还为发布日期和访问级别创建了属性,并在其中填充了随机值作为演示。
from rdflib import Graph, RDF, RDFS, Namespace, URIRef, Literal
from rdflib.namespace import SKOS, XSD
import pandas as pd
import urllib.parse
import random
from datetime import datetime, timedelta
# Create a new RDF graph
g = Graph()
# Define namespaces
schema = Namespace('http://schema.org/')
ex = Namespace('http://example.org/')
prefixes = {
'schema': schema,
'ex': ex,
'skos': SKOS,
'xsd': XSD
}
for p, ns in prefixes.items():
g.bind(p, ns)
# Define classes and properties
Article = URIRef(ex.Article)
MeSHTerm = URIRef(ex.MeSHTerm)
g.add((Article, RDF.type, RDFS.Class))
g.add((MeSHTerm, RDF.type, RDFS.Class))
title = URIRef(schema.name)
abstract = URIRef(schema.description)
date_published = URIRef(schema.datePublished)
access = URIRef(ex.access)
g.add((title, RDF.type, RDF.Property))
g.add((abstract, RDF.type, RDF.Property))
g.add((date_published, RDF.type, RDF.Property))
g.add((access, RDF.type, RDF.Property))
# Function to clean and parse MeSH terms
def parse_mesh_terms(mesh_list):
if pd.isna(mesh_list):
return []
return [term.strip().replace(' ', '_') for term in mesh_list.strip("[]'").split(',')]
# Function to create a valid URI
def create_valid_uri(base_uri, text):
if pd.isna(text):
return None
sanitized_text = urllib.parse.quote(text.strip().replace(' ', '_').replace('"', '').replace('<', '').replace('>', '').replace("'", "_"))
return URIRef(f"{base_uri}/{sanitized_text}")
# Function to generate a random date within the last 5 years
def generate_random_date():
start_date = datetime.now() - timedelta(days=5*365)
random_days = random.randint(0, 5*365)
return start_date + timedelta(days=random_days)
# Function to generate a random access value between 1 and 10
def generate_random_access():
return random.randint(1, 10)
# Load your DataFrame here
# df = pd.read_csv('your_data.csv')
# Loop through each row in the DataFrame and create RDF triples
for index, row in df.iterrows():
article_uri = create_valid_uri("http://example.org/article", row['Title'])
if article_uri is None:
continue
# Add Article instance
g.add((article_uri, RDF.type, Article))
g.add((article_uri, title, Literal(row['Title'], datatype=XSD.string)))
g.add((article_uri, abstract, Literal(row['abstractText'], datatype=XSD.string)))
# Add random datePublished and access
random_date = generate_random_date()
random_access = generate_random_access()
g.add((article_uri, date_published, Literal(random_date.date(), datatype=XSD.date)))
g.add((article_uri, access, Literal(random_access, datatype=XSD.integer)))
# Add MeSH Terms
mesh_terms = parse_mesh_terms(row['meshMajor'])
for term in mesh_terms:
term_uri = create_valid_uri("http://example.org/mesh", term)
if term_uri is None:
continue
# Add MeSH Term instance
g.add((term_uri, RDF.type, MeSHTerm))
g.add((term_uri, RDFS.label, Literal(term.replace('_', ' '), datatype=XSD.string)))
# Link Article to MeSH Term
g.add((article_uri, schema.about, term_uri))
# Serialize the graph to a file (optional)
g.serialize(destination='ontology.ttl', format='turtle')
使用知识图谱进行语义搜索
现在我们可以测试语义搜索了。不过,语义一词在知识图谱中略有不同。在知识图谱中,我们依靠与文档相关的标签以及它们在 MeSH 分类法中的关系来确定语义。例如,一篇文章可能是关于唾液腺肿瘤(唾液腺癌症)的,但仍被标记为口腔肿瘤。
我们不会查询所有标有 “口腔肿瘤 ”的文章,而是会查找比 “口腔肿瘤 ”更窄的概念。MeSH 词汇表包含术语定义,但也包含更宽和更窄等关系。
from SPARQLWrapper import SPARQLWrapper, JSON
def get_concept_triples_for_term(term):
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?subject ?p ?pLabel ?o ?oLabel
FROM <http://id.nlm.nih.gov/mesh>
WHERE {{
?subject rdfs:label "{term}"@en .
?subject ?p ?o .
FILTER(CONTAINS(STR(?p), "concept"))
OPTIONAL {{ ?p rdfs:label ?pLabel . }}
OPTIONAL {{ ?o rdfs:label ?oLabel . }}
}}
"""
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
triples = set() # Using a set to avoid duplicate entries
for result in results["results"]["bindings"]:
obj_label = result.get("oLabel", {}).get("value", "No label")
triples.add(obj_label)
# Add the term itself to the list
triples.add(term)
return list(triples) # Convert back to a list for easier handling
def get_narrower_concepts_for_term(term):
sparql = SPARQLWrapper("https://id.nlm.nih.gov/mesh/sparql")
query = f"""
PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX meshv: <http://id.nlm.nih.gov/mesh/vocab#>
PREFIX mesh: <http://id.nlm.nih.gov/mesh/>
SELECT ?narrowerConcept ?narrowerConceptLabel
WHERE {{
?broaderConcept rdfs:label "{term}"@en .
?narrowerConcept meshv:broaderDescriptor ?broaderConcept .
?narrowerConcept rdfs:label ?narrowerConceptLabel .
}}
"""
sparql.setQuery(query)
sparql.setReturnFormat(JSON)
results = sparql.query().convert()
concepts = set() # Using a set to avoid duplicate entries
for result in results["results"]["bindings"]:
subject_label = result.get("narrowerConceptLabel", {}).get("value", "No label")
concepts.add(subject_label)
return list(concepts) # Convert back to a list for easier handling
def get_all_narrower_concepts(term, depth=2, current_depth=1):
# Create a dictionary to store the terms and their narrower concepts
all_concepts = {}
# Initial fetch for the primary term
narrower_concepts = get_narrower_concepts_for_term(term)
all_concepts[term] = narrower_concepts
# If the current depth is less than the desired depth, fetch narrower concepts recursively
if current_depth < depth:
for concept in narrower_concepts:
# Recursive call to fetch narrower concepts for the current concept
child_concepts = get_all_narrower_concepts(concept, depth, current_depth + 1)
all_concepts.update(child_concepts)
return all_concepts
# Fetch alternative names and narrower concepts
term = "Mouth Neoplasms"
alternative_names = get_concept_triples_for_term(term)
all_concepts = get_all_narrower_concepts(term, depth=2) # Adjust depth as needed
# Output alternative names
print("Alternative names:", alternative_names)
print()
# Output narrower concepts
for broader, narrower in all_concepts.items():
print(f"Broader concept: {broader}")
print(f"Narrower concepts: {narrower}")
print("---")
以下是口腔肿瘤的所有替代名称和狭义概念。
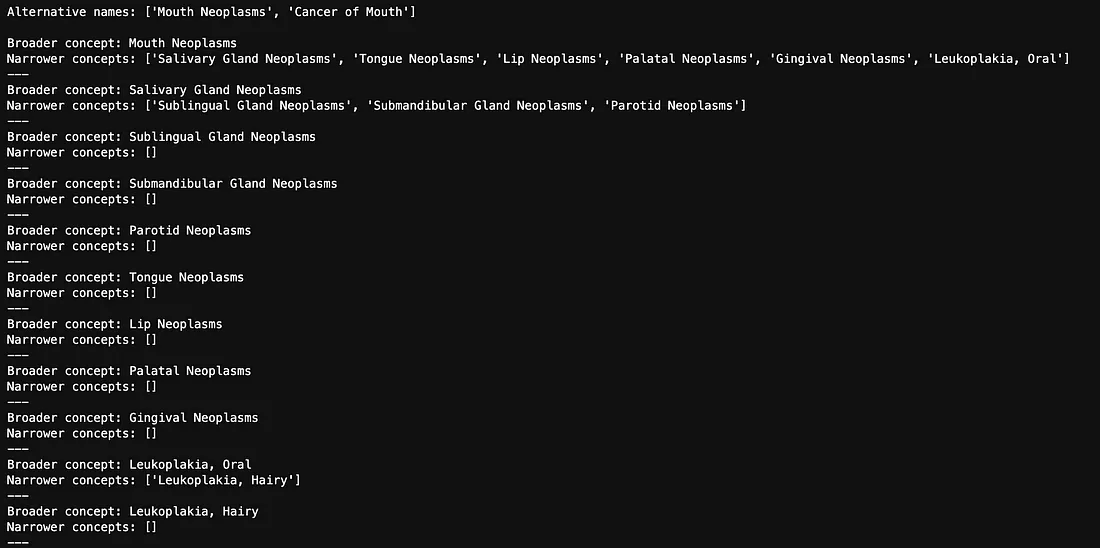
我们将其转化为一个统一的术语列表:
def flatten_concepts(concepts_dict):
flat_list = []
def recurse_terms(term_dict):
for term, narrower_terms in term_dict.items():
flat_list.append(term)
if narrower_terms:
recurse_terms(dict.fromkeys(narrower_terms, [])) # Use an empty dict to recurse
recurse_terms(concepts_dict)
return flat_list
# Flatten the concepts dictionary
flat_list = flatten_concepts(all_concepts)
然后,我们将这些术语转化为 MeSH URI,以便将其纳入 SPARQL 查询:
#Convert the MeSH terms to URI
def convert_to_mesh_uri(term):
formatted_term = term.replace(" ", "_").replace(",", "_").replace("-", "_")
return URIRef(f"http://example.org/mesh/_{formatted_term}_")
# Convert terms to URIs
mesh_terms = [convert_to_mesh_uri(term) for term in flat_list]
然后,我们编写一个 SPARQL 查询,查找所有标有 “口腔肿瘤”、其替代名称 “口腔癌 ”或任何较窄术语的文章:
from rdflib import URIRef
query = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTerm
WHERE {
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
}
"""
# Dictionary to store articles and their associated MeSH terms
article_data = {}
# Run the query for each MeSH term
for mesh_term in mesh_terms:
results = g.query(query, initBindings={'meshTerm': mesh_term})
# Process results
for row in results:
article_uri = row['article']
if article_uri not in article_data:
article_data[article_uri] = {
'title': row['title'],
'abstract': row['abstract'],
'datePublished': row['datePublished'],
'access': row['access'],
'meshTerms': set()
}
# Add the MeSH term to the set for this article
article_data[article_uri]['meshTerms'].add(str(row['meshTerm']))
# Rank articles by the number of matching MeSH terms
ranked_articles = sorted(
article_data.items(),
key=lambda item: len(item[1]['meshTerms']),
reverse=True
)
# Get the top 3 articles
top_3_articles = ranked_articles[:3]
# Output results
for article_uri, data in top_3_articles:
print(f"Title: {data['title']}")
print("MeSH Terms:")
for mesh_term in data['meshTerms']:
print(f" - {mesh_term}")
print()
返回的文章是
- 第 2 条(来自上文): “小唾液腺源性肌上皮瘤。光镜和电子显微镜研究"。
- 第 4 条(引自上文): “针对吸烟者进行口腔恶性病变筛查的可行性研究"。
- 第6篇:“口腔鳞状细胞癌中胚胎致死性异常视觉样蛋白HuR的表达与环氧化酶-2之间的关联”。这篇文章介绍了一项研究,该研究旨在确定一种名为 HuR 的蛋白质的存在是否与较高水平的环氧合酶-2 有关,而环氧合酶-2 在癌症发展和癌细胞扩散中发挥着作用。具体来说,这项研究的重点是口腔鳞状细胞癌(一种口腔癌)。
这些结果与我们从载体数据库中得到的结果并无二致。每篇文章都与口腔肿瘤有关。知识图谱方法的好处在于我们可以获得可解释性--我们清楚地知道为什么会选择这些文章。第 2 篇文章的标签是 “牙龈肿瘤 ”和 “唾液腺肿瘤”。第 4 条和第 6 条的标签都是 “口腔肿瘤”。由于第 2 条被标记了 2 个与我们的搜索条件相匹配的术语,因此排名最高。
使用知识图谱进行相似性搜索
与使用向量空间查找相似文章相比,我们可以依靠与文章相关的标签。使用标签进行相似性搜索有多种方法,但在本例中,我将使用一种常用的方法: 雅卡德相似度。我们将再次使用牙龈文章来比较不同的方法。
from rdflib import Graph, URIRef
from rdflib.namespace import RDF, RDFS, Namespace, SKOS
import urllib.parse
# Define namespaces
schema = Namespace('http://schema.org/')
ex = Namespace('http://example.org/')
rdfs = Namespace('http://www.w3.org/2000/01/rdf-schema#')
# Function to calculate Jaccard similarity and return overlapping terms
def jaccard_similarity(set1, set2):
intersection = set1.intersection(set2)
union = set1.union(set2)
similarity = len(intersection) / len(union) if len(union) != 0 else 0
return similarity, intersection
# Load the RDF graph
g = Graph()
g.parse('ontology.ttl', format='turtle')
def get_article_uri(title):
# Convert the title to a URI-safe string
safe_title = urllib.parse.quote(title.replace(" ", "_"))
return URIRef(f"http://example.org/article/{safe_title}")
def get_mesh_terms(article_uri):
query = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?meshTerm
WHERE {
?article schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
FILTER (?article = <""" + str(article_uri) + """>)
}
"""
results = g.query(query)
mesh_terms = {str(row['meshTerm']) for row in results}
return mesh_terms
def find_similar_articles(title):
article_uri = get_article_uri(title)
mesh_terms_given_article = get_mesh_terms(article_uri)
# Query all articles and their MeSH terms
query = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?article ?meshTerm
WHERE {
?article a ex:Article ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
}
"""
results = g.query(query)
mesh_terms_other_articles = {}
for row in results:
article = str(row['article'])
mesh_term = str(row['meshTerm'])
if article not in mesh_terms_other_articles:
mesh_terms_other_articles[article] = set()
mesh_terms_other_articles[article].add(mesh_term)
# Calculate Jaccard similarity
similarities = {}
overlapping_terms = {}
for article, mesh_terms in mesh_terms_other_articles.items():
if article != str(article_uri):
similarity, overlap = jaccard_similarity(mesh_terms_given_article, mesh_terms)
similarities[article] = similarity
overlapping_terms[article] = overlap
# Sort by similarity and get top 5
top_similar_articles = sorted(similarities.items(), key=lambda x: x[1], reverse=True)[:15]
# Print results
print(f"Top 15 articles similar to '{title}':")
for article, similarity in top_similar_articles:
print(f"Article URI: {article}")
print(f"Jaccard Similarity: {similarity:.4f}")
print(f"Overlapping MeSH Terms: {overlapping_terms[article]}")
print()
# Example usage
article_title = "Gingival metastasis as first sign of multiorgan dissemination of epithelioid malignant mesothelioma."
find_similar_articles(article_title)
搜索结果如下。由于我们再次搜索的是 “牙龈 ”这篇文章,因此它是相似度最高的文章,这也是我们所期望的。其他结果如下:
- 文章7:"侧阔肌钙化性肌腱炎。三个病例的报告"。这篇文章是关于大腿外侧肌(大腿上的一块肌肉)的钙化性腱鞘炎(钙沉积在肌腱中)。这与口腔肿瘤无关。
- 重叠术语: X射线断层扫描,老年人,男性,人类,X射线计算
- 文章8:“对于出现前列腺特异性抗原水平的前列腺癌患者,雄激素剥夺疗法的最佳持续时间是多久”。这篇文章是关于前列腺癌患者应该接受多长时间的特定治疗(雄激素剥夺疗法)。这是关于癌症的治疗(放疗),但不是口腔癌。
- 重叠术语: 放疗 老年 男性 人类 辅助治疗
- 文章9:CT扫描大脑半球不对称:失语症康复的预测因素。本文介绍大脑左右两侧的差异(大脑半球不对称)如何预测中风后失语症患者的恢复情况。
- 重叠术语: 断层扫描,年龄,男性,人类,X 射线计算
这种方法的优点在于,由于我们在这里采用的是计算相似度的方法,因此我们可以看到其他文章为什么相似--我们可以准确地看到哪些术语是重叠的,即哪些术语在 Gingival 文章和每篇比较文章中是共同的。
可解释性的缺点在于,根据之前的结果,我们可以发现这些文章似乎并不是最相似的文章。它们都有三个共同的术语(年龄、男性和人类),而这三个术语的相关性可能远不及治疗方案或口腔肿瘤。你可以根据术语在整个语料库中的流行程度(术语频率-反向文档频率(TF-IDF))来重新计算权重,这样可能会改善结果。在进行相似性计算时,你还可以选择与你最相关的标签术语,以便对结果进行更多控制。
对知识图谱中的术语使用 Jaccard 相似性来计算相似性的最大缺点是计算量大,计算一次大概需要 30 分钟。
使用知识图谱进行 RAG
我们也可以只使用知识图谱进行检索部分的 RAG 计算。我们已经有一个关于口腔肿瘤的文章列表,作为上述语义搜索的结果保存下来。要实现 RAG,我们只需将这些文章发送给 LLM,并要求它对结果进行总结。
首先,我们将每篇文章的标题和摘要合并成一大段文本,称为 combined_text:
# Function to combine titles and abstracts
def combine_abstracts(top_3_articles):
combined_text = "".join(
[f"Title: {data['title']} Abstract: {data['abstract']}" for article_uri, data in top_3_articles]
)
return combined_text
# Combine abstracts from the top 3 articles
combined_text = combine_abstracts(top_3_articles)
print(combined_text)
然后,我们建立一个客户端,这样就可以直接将文本发送给LLM:
import openai
# Set up your OpenAI API key
api_key = "YOUR API KEY"
openai.api_key = api_key
然后,我们向 LLM 提供背景和提示:
def generate_summary(combined_text):
response = openai.Completion.create(
model="gpt-3.5-turbo-instruct",
prompt=f"Summarize the key information here in bullet points. Make it understandable to someone without a medical degree:\n\n{combined_text}",
max_tokens=1000,
temperature=0.3
)
# Get the raw text output
raw_summary = response.choices[0].text.strip()
# Split the text into lines and clean up whitespace
lines = raw_summary.split('\n')
lines = [line.strip() for line in lines if line.strip()]
# Join the lines back together with actual line breaks
formatted_summary = '\n'.join(lines)
return formatted_summary
# Generate and print the summary
summary = generate_summary(combined_text)
print(summary)
结果如下:
- A 14-year-old boy had a gingival tumor in his anterior maxilla that was removed and studied by light and electron microscopy
- The tumor was made up of myoepithelial cells and appeared to be malignant
- Electron microscopy showed that the tumor originated from a salivary gland
- This is the only confirmed case of a myoepithelioma with features of malignancy
- A feasibility study was conducted to improve early detection of oral cancer and premalignant lesions in a high incidence region
- Tobacco vendors were involved in distributing flyers to invite smokers for free examinations by general practitioners
- 93 patients were included in the study and 27% were referred to a specialist
- 63.6% of those referred actually saw a specialist and 15.3% were confirmed to have a premalignant lesion
- A study found a correlation between increased expression of the protein HuR and the enzyme COX-2 in oral squamous cell carcinoma (OSCC)
- Cytoplasmic HuR expression was associated with COX-2 expression and lymph node and distant metastasis in OSCCs
- Inhibition of HuR expression led to a decrease in COX-2 expression in oral cancer cells.
结果看起来不错,也就是说,这是对语义搜索返回的三篇文章的很好总结。仅使用 KG 的 RAG 应用程序的响应质量取决于 KG 检索相关文档的能力。从本示例中可以看出,如果你的提示足够简单,例如 “总结此处的关键信息”,那么困难的部分就在于检索(为 LLM 提供正确的文章作为上下文),而不是生成响应。
第3步:使用矢量化知识图谱测试数据检索
现在我们要联合起来。我们将为数据库中的每篇文章添加一个 URI,然后在 Weaviate 中创建一个新的集合,将文章名称、摘要、与之相关的 MeSH 术语以及 URI 矢量化。URI 是文章的唯一标识符,也是我们连接回知识图谱的一种方式。
首先,我们在数据中为 URI 添加一个新列:
# Function to create a valid URI
def create_valid_uri(base_uri, text):
if pd.isna(text):
return None
# Encode text to be used in URI
sanitized_text = urllib.parse.quote(text.strip().replace(' ', '_').replace('"', '').replace('<', '').replace('>', '').replace("'", "_"))
return URIRef(f"{base_uri}/{sanitized_text}")
# Add a new column to the DataFrame for the article URIs
df['Article_URI'] = df['Title'].apply(lambda title: create_valid_uri("http://example.org/article", title))
现在,我们为带有附加字段的新集合创建一个新模式:
class_obj = {= {
# Class definition
"class": "articles_with_abstracts_and_URIs",
# Property definitions
"properties": [
{
"name": "title",
"dataType": ["text"],
},
{
"name": "abstractText",
"dataType": ["text"],
},
{
"name": "meshMajor",
"dataType": ["text"],
},
{
"name": "Article_URI",
"dataType": ["text"],
},
],
# Specify a vectorizer
"vectorizer": "text2vec-openai",
# Module settings
"moduleConfig": {
"text2vec-openai": {
"vectorizeClassName": True,
"model": "ada",
"modelVersion": "002",
"type": "text"
},
"qna-openai": {
"model": "gpt-3.5-turbo-instruct"
},
"generative-openai": {
"model": "gpt-3.5-turbo"
}
},
}将该模式推送到矢量数据库:
client.schema.create_class(class_obj)schema.create_class(class_obj)
现在,我们将数据矢量化到新的集合中:
import logging
import numpy as np
# Configure logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s %(levelname)s %(message)s')
# Replace infinity values with NaN and then fill NaN values
df.replace([np.inf, -np.inf], np.nan, inplace=True)
df.fillna('', inplace=True)
# Convert columns to string type
df['Title'] = df['Title'].astype(str)
df['abstractText'] = df['abstractText'].astype(str)
df['meshMajor'] = df['meshMajor'].astype(str)
df['Article_URI'] = df['Article_URI'].astype(str)
# Log the data types
logging.info(f"Title column type: {df['Title'].dtype}")
logging.info(f"abstractText column type: {df['abstractText'].dtype}")
logging.info(f"meshMajor column type: {df['meshMajor'].dtype}")
logging.info(f"Article_URI column type: {df['Article_URI'].dtype}")
with client.batch(
batch_size=10, # Specify batch size
num_workers=2, # Parallelize the process
) as batch:
for index, row in df.iterrows():
try:
question_object = {
"title": row.Title,
"abstractText": row.abstractText,
"meshMajor": row.meshMajor,
"article_URI": row.Article_URI,
}
batch.add_data_object(
question_object,
class_name="articles_with_abstracts_and_URIs",
uuid=generate_uuid5(question_object)
)
except Exception as e:
logging.error(f"Error processing row {index}: {e}")
利用矢量化知识图谱进行语义搜索
现在,我们可以在矢量数据库上进行语义搜索,就像以前一样,但结果更具可解释性和可控性。
response = (
client.query
.get("articles_with_abstracts_and_URIs", ["title","abstractText","meshMajor","article_URI"])get("articles_with_abstracts_and_URIs", ["title","abstractText","meshMajor","article_URI"])
.with_additional(["id"])
.with_near_text({"concepts": ["mouth neoplasms"]})
.with_limit(10)
.do()
)
print(json.dumps(response, indent=4))
结果如下
- 第1篇:“牙龈转移是上皮样恶性间皮瘤多器官播散的首发症状”。
- 文章10:“血管中心性面中部淋巴瘤是一名老年人的诊断难题”。这篇文章讲述了如何对一名患有鼻癌的男子进行具有挑战性的诊断。
- 文章11:“下颌骨假癌增生”。这篇文章对我来说很难解读,但我相信它讲述的是假癌增生看起来像癌症(因此名字中出现了 pseuo),但其实并不是癌症。虽然这篇文章似乎是关于下颌骨的,但它被标记为 MeSH 术语 “口腔肿瘤”。
很难说这些结果比单独的 KG 或载体数据库更好还是更差。从理论上讲,结果应该更好,因为与每篇文章相关的 MeSH 术语现在都与文章一起被矢量化了。不过,我们并没有真正将知识图谱矢量化。例如,MeSH 术语之间的关系并不在矢量数据库中。
将 MeSH 术语矢量化的好处是可以立即进行解释--例如,第 11 条也标记为 “口腔肿瘤”。不过,将矢量数据库与知识图谱连接起来的真正好处是,我们可以从知识图谱中应用我们想要的任何过滤器。还记得我们之前是如何在数据中添加发布日期字段的吗?现在我们可以对此进行筛选。假设我们想查找 2020 年 5 月 1 日之后发表的有关口腔肿瘤的文章:
from rdflib import Graph, Namespace, URIRef, Literal
from rdflib.namespace import RDF, RDFS, XSD
# Define namespaces
schema = Namespace('http://schema.org/')
ex = Namespace('http://example.org/')
rdfs = Namespace('http://www.w3.org/2000/01/rdf-schema#')
xsd = Namespace('http://www.w3.org/2001/XMLSchema#')
def get_articles_after_date(graph, article_uris, date_cutoff):
# Create a dictionary to store results for each URI
results_dict = {}
# Define the SPARQL query using a list of article URIs and a date filter
uris_str = " ".join(f"<{uri}>" for uri in article_uris)
query = f"""
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
SELECT ?article ?title ?datePublished
WHERE {{
VALUES ?article {{ {uris_str} }}
?article a ex:Article ;
schema:name ?title ;
schema:datePublished ?datePublished .
FILTER (?datePublished > "{date_cutoff}"^^xsd:date)
}}
"""
# Execute the query
results = graph.query(query)
# Extract the details for each article
for row in results:
article_uri = str(row['article'])
results_dict[article_uri] = {
'title': str(row['title']),
'date_published': str(row['datePublished'])
}
return results_dict
date_cutoff = "2023-01-01"
articles_after_date = get_articles_after_date(g, article_uris, date_cutoff)
# Output the results
for uri, details in articles_after_date.items():
print(f"Article URI: {uri}")
print(f"Title: {details['title']}")
print(f"Date Published: {details['date_published']}")
print()
最初的查询返回了十条结果(我们设定的最大值为十条),但其中只有六条是在 2023 年 1 月 1 日之后发布的。请看下面的结果:
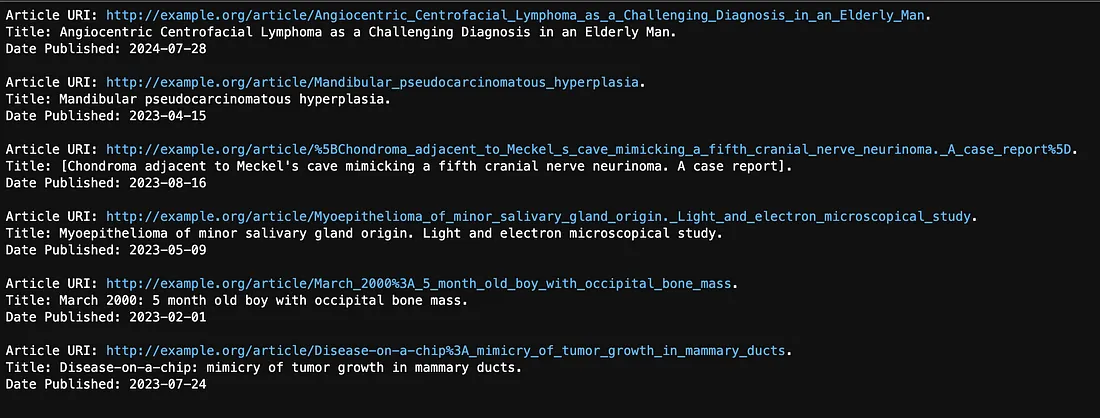
使用矢量化知识图谱进行相似性搜索
我们可以像之前在牙龈文章(第 1 条)中所做的那样,在这个新集合上运行相似性搜索:
response = (
client.query
.get("articles_with_abstracts_and_URIs", ["title","abstractText","meshMajor","article_URI"])get("articles_with_abstracts_and_URIs", ["title","abstractText","meshMajor","article_URI"])
.with_near_object({
"id": "37b695c4-5b80-5f44-a710-e84abb46bc22"
})
.with_limit(50)
.with_additional(["distance"])
.do()
)
print(json.dumps(response, indent=2))
结果如下:
- 第 3 条:"下颌骨转移性神经母细胞瘤。病例报告"。
- 第 4 条:“针对烟草使用者进行口腔恶性病变筛查的可行性研究”。
- 文章12:“伪装成间质性肺病的弥漫性肺内恶性间皮瘤:间皮瘤的一种独特变异”。这篇文章讲述了五名男性间皮瘤患者的病情,这种间皮瘤看起来很像另一种肺病:间质性肺病。
由于我们对 MeSH 标记进行了矢量化,因此我们可以看到与每篇文章相关的标记。其中有些文章虽然在某些方面可能相似,但却与口腔肿瘤无关。假设我们想查找与我们的牙龈文章类似,但具体涉及口腔肿瘤的文章。现在,我们可以将之前进行的 SPARQL 过滤与知识图谱结合起来,对这些结果进行处理。
我们已经保存了 “口腔肿瘤 ”的同义词和狭义概念的 MeSH URI,但还需要矢量搜索返回的 50 篇文章的 URI:
# Assuming response is the data structure with your articles
article_uris = [URIRef(article["article_URI"]) for article in response["data"]["Get"]["Articles_with_abstracts_and_URIs"]]
现在,我们可以根据标签对结果进行排序,就像之前使用知识图谱进行语义搜索一样。
from rdflib import URIRef
# Constructing the SPARQL query with a FILTER for the article URIs
query = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTerm
WHERE {
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
# Filter to include only articles from the list of URIs
FILTER (?article IN (%s))
}
"""
# Convert the list of URIRefs into a string suitable for SPARQL
article_uris_string = ", ".join([f"<{str(uri)}>" for uri in article_uris])
# Insert the article URIs into the query
query = query % article_uris_string
# Dictionary to store articles and their associated MeSH terms
article_data = {}
# Run the query for each MeSH term
for mesh_term in mesh_terms:
results = g.query(query, initBindings={'meshTerm': mesh_term})
# Process results
for row in results:
article_uri = row['article']
if article_uri not in article_data:
article_data[article_uri] = {
'title': row['title'],
'abstract': row['abstract'],
'datePublished': row['datePublished'],
'access': row['access'],
'meshTerms': set()
}
# Add the MeSH term to the set for this article
article_data[article_uri]['meshTerms'].add(str(row['meshTerm']))
# Rank articles by the number of matching MeSH terms
ranked_articles = sorted(
article_data.items(),
key=lambda item: len(item[1]['meshTerms']),
reverse=True
)
# Output results
for article_uri, data in ranked_articles:
print(f"Title: {data['title']}")
print(f"Abstract: {data['abstract']}")
print("MeSH Terms:")
for mesh_term in data['meshTerms']:
print(f" - {mesh_term}")
print()
在矢量数据库最初返回的 50 篇文章中,只有 5 篇标注了口腔肿瘤或相关概念。
- 文章 2:"小唾液腺源性肌上皮瘤。光镜和电子显微镜研究"。标签:: 牙龈肿瘤、唾液腺肿瘤
- 文章4:“针对吸烟者进行口腔恶性病变筛查的可行性研究”。标签: 口腔肿瘤 口腔肿瘤
- 文章13:“源于龈沟的表皮样癌”。本文描述了一例牙龈癌(牙龈肿瘤)病例。标签: 牙龈肿瘤
- 文章1:“牙龈转移是上皮样恶性间皮瘤多器官播散的首发症状”。标签: 上皮样恶性间皮瘤 牙龈肿瘤
- 文章14:"腮腺结节转移: CT和MR成像发现"。本文介绍腮腺(主要唾液腺)中的肿瘤。标签: 腮腺肿瘤
最后,假设我们想向用户推荐这些类似的文章,但我们只想推荐该用户可以访问的文章。假设我们知道该用户只能访问权限级别为 3、5 和 7 的文章。我们可以使用类似的 SPARQL 查询在知识图谱中应用过滤器:
from rdflib import Graph, Namespace, URIRef, Literal
from rdflib.namespace import RDF, RDFS, XSD, SKOS
# Assuming your RDF graph (g) is already loaded
# Define namespaces
schema = Namespace('http://schema.org/')
ex = Namespace('http://example.org/')
rdfs = Namespace('http://www.w3.org/2000/01/rdf-schema#')
def filter_articles_by_access(graph, article_uris, access_values):
# Construct the SPARQL query with a dynamic VALUES clause
uris_str = " ".join(f"<{uri}>" for uri in article_uris)
query = f"""
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTermLabel
WHERE {{
VALUES ?article {{ {uris_str} }}
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm rdfs:label ?meshTermLabel .
FILTER (?access IN ({", ".join(map(str, access_values))}))
}}
"""
# Execute the query
results = graph.query(query)
# Extract the details for each article
results_dict = {}
for row in results:
article_uri = str(row['article'])
if article_uri not in results_dict:
results_dict[article_uri] = {
'title': str(row['title']),
'abstract': str(row['abstract']),
'date_published': str(row['datePublished']),
'access': str(row['access']),
'mesh_terms': []
}
results_dict[article_uri]['mesh_terms'].append(str(row['meshTermLabel']))
return results_dict
access_values = [3,5,7]
filtered_articles = filter_articles_by_access(g, ranked_article_uris, access_values)
# Output the results
for uri, details in filtered_articles.items():
print(f"Article URI: {uri}")
print(f"Title: {details['title']}")
print(f"Abstract: {details['abstract']}")
print(f"Date Published: {details['date_published']}")
print(f"Access: {details['access']}")
print()
有一篇文章用户无法访问。其余四篇文章是:
- 文章2:"小唾液腺源性肌上皮瘤。光镜和电子显微镜研究"。标签:: 牙龈肿瘤, 唾液腺肿瘤. 访问级别:5
- 文章4:“针对烟草使用者进行口腔恶性病变筛查的可行性研究”。标签:: 口腔肿瘤。访问级别:7
- 文章1:“牙龈转移是上皮样恶性间皮瘤多器官播散的首发症状”。Tagged with: 牙龈肿瘤。访问级别:3
- 文章14:"腮腺结节转移: CT和MR成像结果"。本文介绍腮腺(主要唾液腺)中的肿瘤。标签: 腮腺肿瘤。访问级别:3
带有矢量化知识图谱的 RAG
最后,让我们来看看将矢量数据库与知识图谱相结合后,RAG 是如何工作的。提醒一下,你可以直接针对向量数据库运行 RAG,并将其发送到 LLM,以获得生成的响应:
response = (
client.query
.get("Articles_with_abstracts_and_URIs", ["title", "abstractText",'article_URI','meshMajor'])get("Articles_with_abstracts_and_URIs", ["title", "abstractText",'article_URI','meshMajor'])
.with_near_text({"concepts": ["therapies for mouth neoplasms"]})
.with_limit(3)
.with_generate(grouped_task="Summarize the key information here in bullet points. Make it understandable to someone without a medical degree.")
.do()
)
print(response["data"]["Get"]["Articles_with_abstracts_and_URIs"][0]["_additional"]["generate"]["groupedResult"])
在这个例子中,我使用的搜索词是 “口腔肿瘤的治疗方法”,同样的提示是 "用要点概括这里的关键信息。让没有医学学位的人也能理解"。“我们只返回前三篇文章来生成此回复。以下是结果:
- Metastatic malignant mesothelioma to the oral cavity is rare, with an average survival rate of 9-12 months.
- Neoadjuvant chemotherapy and radical pleurectomy decortication followed by radiotherapy were used in 13 patients from August 2012 to September 2013.
- In January 2014, 11 patients were still alive with a median survival of 11 months, while 8 patients had a recurrence and 2 patients died at 8 and 9 months after surgery.
- A 68-year-old man had a gingival mass that turned out to be a metastatic deposit of malignant mesothelioma, leading to multiorgan recurrence.
- Biopsy is important for new growing lesions, even in uncommon sites, when there is a history of mesothelioma.
- Neoadjuvant radiochemotherapy for locally advanced rectal carcinoma can be effective, but some patients may not respond well.
- Genetic alterations may be associated with sensitivity or resistance to neoadjuvant therapy in rectal cancer.
- Losses of chromosomes 1p, 8p, 17p, and 18q, and gains of 1q and 13q were found in rectal cancer tumors.
- Alterations in specific chromosomal regions were associated with the response to neoadjuvant therapy.
- The cytogenetic profile of tumor cells may influence the response to radiochemotherapy in rectal cancer.
- Intensity-modulated radiation therapy for nasopharyngeal carcinoma achieved good long-term outcomes in terms of local control and overall survival.
- Acute toxicities included mucositis, dermatitis, and xerostomia, with most patients experiencing Grade 0-2 toxicities.
- Late toxicity mainly included xerostomia, which improved over time.
- Distant metastasis remained the main cause of treatment failure, highlighting the need for more effective systemic therapy.
作为测试,我们可以看到到底选择了哪三篇文章:
# Extract article URIs
article_uris = [article["article_URI"] for article in response["data"]["Get"]["Articles_with_abstracts_and_URIs"]]
# Function to filter the response for only the given URIs
def filter_articles_by_uri(response, article_uris):
filtered_articles = []
articles = response['data']['Get']['Articles_with_abstracts_and_URIs']
for article in articles:
if article['article_URI'] in article_uris:
filtered_articles.append(article)
return filtered_articles
# Filter the response
filtered_articles = filter_articles_by_uri(response, article_uris)
# Output the filtered articles
print("Filtered articles:")
for article in filtered_articles:
print(f"Title: {article['title']}")
print(f"URI: {article['article_URI']}")
print(f"Abstract: {article['abstractText']}")
print(f"MeshMajor: {article['meshMajor']}")
print("---")
有趣的是,第一篇文章是关于牙龈肿瘤的,而牙龈肿瘤是口腔肿瘤的一个分支,但第二篇文章是关于直肠癌的,第三篇文章是关于鼻咽癌的。它们都是关于癌症的疗法,只是与我搜索的癌症种类不同。令人担忧的是,搜索提示是 “口腔肿瘤的疗法”,而搜索结果却包含了关于其他类型癌症疗法的信息。这就是有时所说的 “语境中毒”--在提示中注入了不相关或误导性的信息,从而导致法律硕士做出误导性的回答。
我们可以使用 KG 来解决上下文中毒问题。下面的图表展示了向量数据库和 KG 如何协同工作,以更好地实施 RAG:
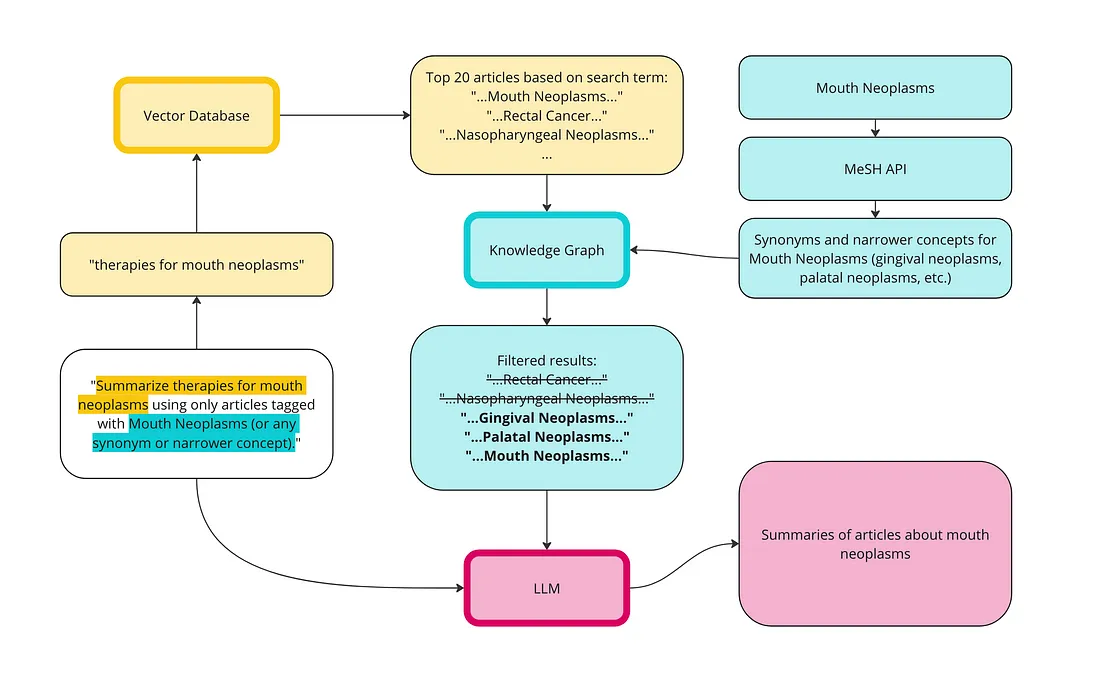
首先,我们使用相同的提示在矢量数据库中进行语义搜索:口腔癌疗法。这次我将限制提高到了 20 篇,因为我们要过滤掉一些文章。
response = (
client.query
.get("articles_with_abstracts_and_URIs", ["title", "abstractText", "meshMajor", "article_URI"])"articles_with_abstracts_and_URIs", ["title", "abstractText", "meshMajor", "article_URI"])
.with_additional(["id"])
.with_near_text({"concepts": ["therapies for mouth neoplasms"]})
.with_limit(20)
.do()
)
# Extract article URIs
article_uris = [article["article_URI"] for article in response["data"]["Get"]["Articles_with_abstracts_and_URIs"]]
# Print the extracted article URIs
print("Extracted article URIs:")
for uri in article_uris:
print(uri)
接下来,我们使用与之前相同的排序技术,使用与口腔肿瘤相关的概念:
from rdflib import URIRef
# Constructing the SPARQL query with a FILTER for the article URIs
query = """
PREFIX schema: <http://schema.org/>
PREFIX ex: <http://example.org/>
SELECT ?article ?title ?abstract ?datePublished ?access ?meshTerm
WHERE {
?article a ex:Article ;
schema:name ?title ;
schema:description ?abstract ;
schema:datePublished ?datePublished ;
ex:access ?access ;
schema:about ?meshTerm .
?meshTerm a ex:MeSHTerm .
# Filter to include only articles from the list of URIs
FILTER (?article IN (%s))
}
"""
# Convert the list of URIRefs into a string suitable for SPARQL
article_uris_string = ", ".join([f"<{str(uri)}>" for uri in article_uris])
# Insert the article URIs into the query
query = query % article_uris_string
# Dictionary to store articles and their associated MeSH terms
article_data = {}
# Run the query for each MeSH term
for mesh_term in mesh_terms:
results = g.query(query, initBindings={'meshTerm': mesh_term})
# Process results
for row in results:
article_uri = row['article']
if article_uri not in article_data:
article_data[article_uri] = {
'title': row['title'],
'abstract': row['abstract'],
'datePublished': row['datePublished'],
'access': row['access'],
'meshTerms': set()
}
# Add the MeSH term to the set for this article
article_data[article_uri]['meshTerms'].add(str(row['meshTerm']))
# Rank articles by the number of matching MeSH terms
ranked_articles = sorted(
article_data.items(),
key=lambda item: len(item[1]['meshTerms']),
reverse=True
)
# Output results
for article_uri, data in ranked_articles:
print(f"Title: {data['title']}")
print(f"Abstract: {data['abstract']}")
print("MeSH Terms:")
for mesh_term in data['meshTerms']:
print(f" - {mesh_term}")
print()
只有三篇文章被标记为口腔肿瘤术语之一:
- 文章4:“针对烟草使用者进行口腔恶性病变筛查的可行性研究”。标签: 口腔肿瘤。
- 第 15 条:“光纤维蛋白介导的光动力疗法对大鼠腭粘膜化学诱导的癌前病变和鳞状细胞癌的治疗”。这篇文章介绍了在大鼠身上测试的一种治疗腭癌的实验性癌症疗法(光动力疗法)。标签: 腭部肿瘤。
- 文章1:“牙龈转移是上皮样恶性间皮瘤多器官播散的首发症状”。标签: 上皮样恶性间皮瘤 牙龈肿瘤。
让我们把这些文章发送到 LLM,看看结果是否有所改善:
# Filter the response
filtered_articles = filter_articles_by_uri(response, matching_articles)
# Function to combine titles and abstracts into one chunk of text
def combine_abstracts(filtered_articles):
combined_text = "\n\n".join(
[f"Title: {article['title']}\nAbstract: {article['abstractText']}" for article in filtered_articles]
)
return combined_text
# Combine abstracts from the filtered articles
combined_text = combine_abstracts(filtered_articles)
# Generate and print the summary
summary = generate_summary(combined_text)
print(summary)
这里是结果:
- Oral cavity cancer is common and often not detected until it is advanced
- A feasibility study was conducted to improve early detection of oral cancer and premalignant lesions in a high-risk region
- Tobacco vendors were involved in distributing flyers to smokers for free examinations by general practitioners
- 93 patients were included in the study, with 27% being referred to a specialist
- 63.6% of referred patients actually saw a specialist, with 15.3% being diagnosed with a premalignant lesion
- Photodynamic therapy (PDT) was studied as an experimental cancer therapy in rats with chemically-induced premalignant lesions and squamous cell carcinoma of the palatal mucosa
- PDT was performed using Photofrin and two different activation wavelengths, with better results seen in the 514.5 nm group
- Gingival metastasis from malignant mesothelioma is extremely rare, with a low survival rate
- A case study showed a patient with a gingival mass as the first sign of multiorgan recurrence of malignant mesothelioma, highlighting the importance of biopsy for all new lesions, even in uncommon anatomical sites.
我们肯定能看到进步--这些结果不是关于直肠癌或鼻咽肿瘤的。这看起来是对所选三篇关于口腔肿瘤疗法的文章相对准确的总结
结论
总的来说,矢量数据库非常适合快速启动和运行搜索、相似性(推荐)和 RAG 应用程序。所需的开销很少。如果你的非结构化数据与结构化数据相关联,就像本例中的期刊论文一样,效果会很好。如果数据集中没有文章摘要,效果就不会那么好。